Research
I develop Machine Learning methods to tackle large-scale Neuroscience problems probabilistically.
Methodology: Plate-Amortized Variational Inference (PAVI)
PAVI is a method to perform inference in large-scale problems. Experimenters frequently hypothesize generative models to explain some observed data. The goal is to recover the latent parameters that could have yielded the observed data through the explanatory model. This is called inference. Because there may be some noise in the system or the generative process is random, the parameters often cannot be inferred unequivocally. The experimenter thus wishes to recover a probabilistic distribution of the parameters. What is the probability that each set of parameters generated the observed data through the model? To give a concrete Neuroscience example: given some MRI imaging of a patient’s brain, how can we guess the size of the neurons in a given part of the brain?
One way to tackle inference is through optimization. We can define parametric distributions and fit those to the unknown distribution of parameters that could explain the observed data. Those methods are coined Variational Inference (VI). VI is an attractive way to tackle inference because it is based on optimization, so it can leverage recent advances in the Machine Learning community. Nevertheless, VI can struggle when tackling large problems. Neuroscience typically features such large scales. For instance, a single brain can feature millions of voxels (volumetric pixels). The goal of PAVI is thus to scale VI to very large problems, featuring hundreds of millions of inferred parameters.
PAVI shares the learning across repeated structures in the problem to scale up inference. As an illustration, having inferred the size of the neurons in a given part of the brain, PAVI does not try to solve similar problems from scratch but does so similarly. In this case: inferring the size of the neurons in different brain parts. Because it shares learning, PAVI can perform inference up to a thousand times faster than off-the-shelf methods. This reduces the training time from weeks to hours and unlocks inference in large-scale regimes. The following sections describe ongoing Neuroscience projects that exploit PAVI.
Check out ![]() the PAVI preprint for more information.
the PAVI preprint for more information.
PAVI is the methodological continuation of ![]() the ADAVI paper published at ICLR 2022.
the ADAVI paper published at ICLR 2022.
Application: full cortex probabilistic parcellation for a cohort of 1,000 subjects
A parcellation clusters brain regions into different connectivity networks: fingerprints describing co-activation with the rest of the brain, as measured using functional Magnetic Resonance Imaging (fMRI). The core idea is that brain regions that co-activate work together towards a given brain function: vision, limb control, etc… A parcellation thus amounts to a functional cartography of the brain.
However, the brains of two different subjects can differ a lot from one another. Furthermore, the information we have to infer the function of a given brain location for a given subject is scarce. Hence, representing the cartographies of different subjects as only probabilistically known is helpful. See below the result for two subjects amongst a population of a thousand. Using PAVI, we shared the learning across brain regions and subjects and got those maps under 5 hours of computation time for a parameter space of 400 million! The maps represent as colors the different networks in the brain and the primarily associated function. When the coloring softens, we cannot associate the corresponding region unequivocally with a network:
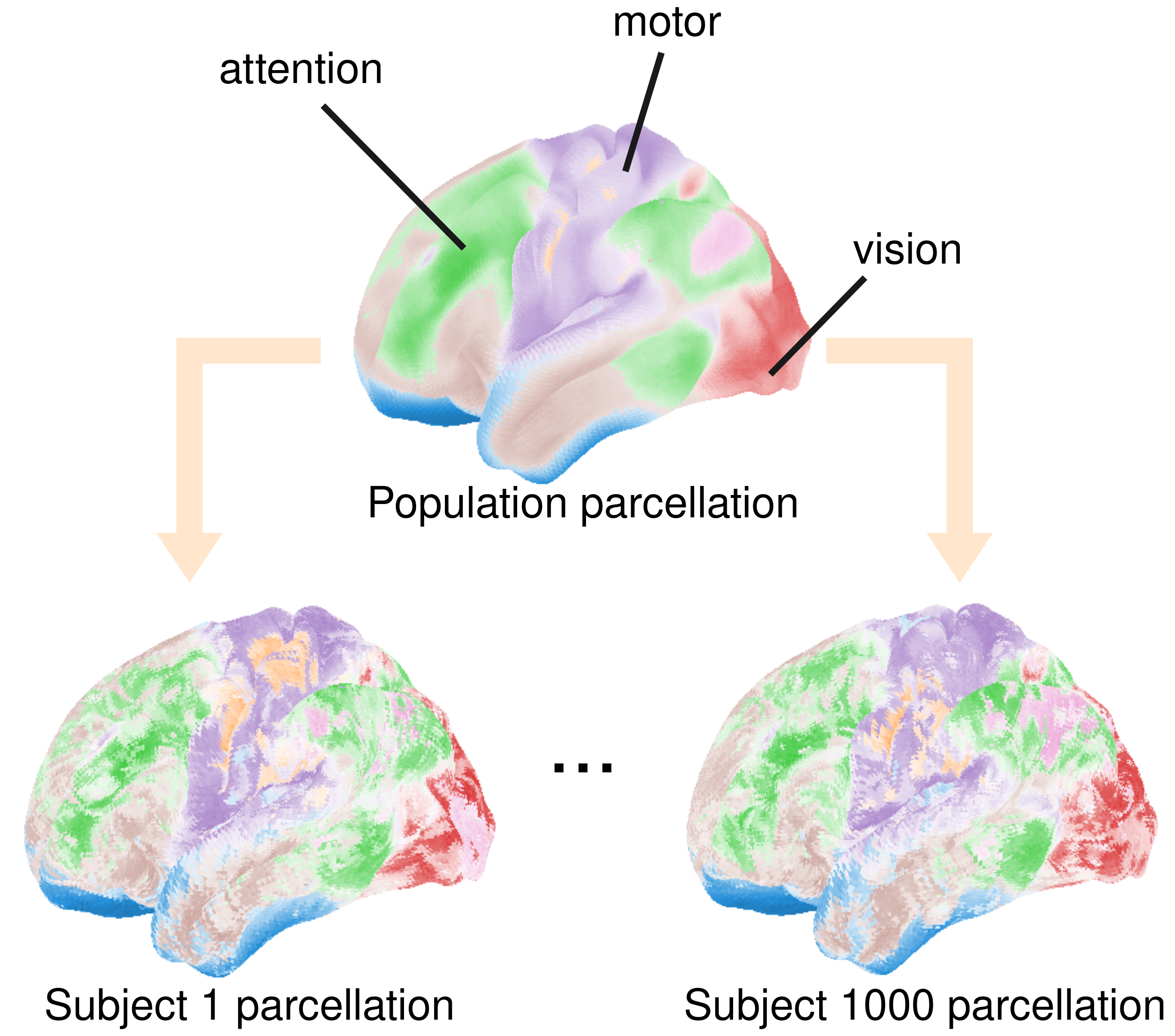
Sources:
Ru Kong, Jingwei Li, Csaba Orban, Mert Rory Sabuncu, Hesheng Liu, Alexander Schaefer, Nanbo Sun, Xi-Nian Zuo, Avram J. Holmes, Simon B. Eickhoff, and B. T. Thomas Yeo. Spatial topography of individual-specific cortical networks predicts human cognition, personality, and emotion. Cerebral cortex, 29 6:2533–2551, 2019.
B. T. Thomas Yeo, Fenna M. Krienen, Jorge Sepulcre, Mert R. Sabuncu, Danial Lashkari, Marisa Hollinshead, Joshua L. Roffman, Jordan W. Smoller, Lilla Zöllei, Jonathan R. Polimeni, Bruce Fischl, Hesheng Liu, and Randy L. Buckner. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. Journal of Neurophysiology, 106(3):1125–1165, 2011.
Application: inferring the cytoarchitecture of the brain in vivo
The cytoarchitecture of the brain describes the microscopic structure of the cells that compose it —for instance, the size of the neurons in a given tissue or the orientation of their axons. Traditionally, to uncover this information, scientists needed to dissect the brain of a dead individual. Nowadays, diffusion Magnetic Resonance Imaging (dMRI) allows us to infer this structure from a brain scan performed in vivo!
However, the link between the cell structure and the measured magnetic field is very complex. Different combinations of cytoarchitectural parameters can yield the same signal. Dealing with this inference in a probabilistic manner is thus valuable. Currently, we study brain regions independently, which can result in significant uncertainty. In this project, I hypothesize some structure linking different brain regions to reduce this uncertainty. For instance, could we hypothesize that the orientation of the neurons smoothly varies across the brain? Below is an example map of the diffusivity of water molecules across brain tissues that helps distinguish the grey matter (at the surface of the cortex) from the white matter (below the surface):
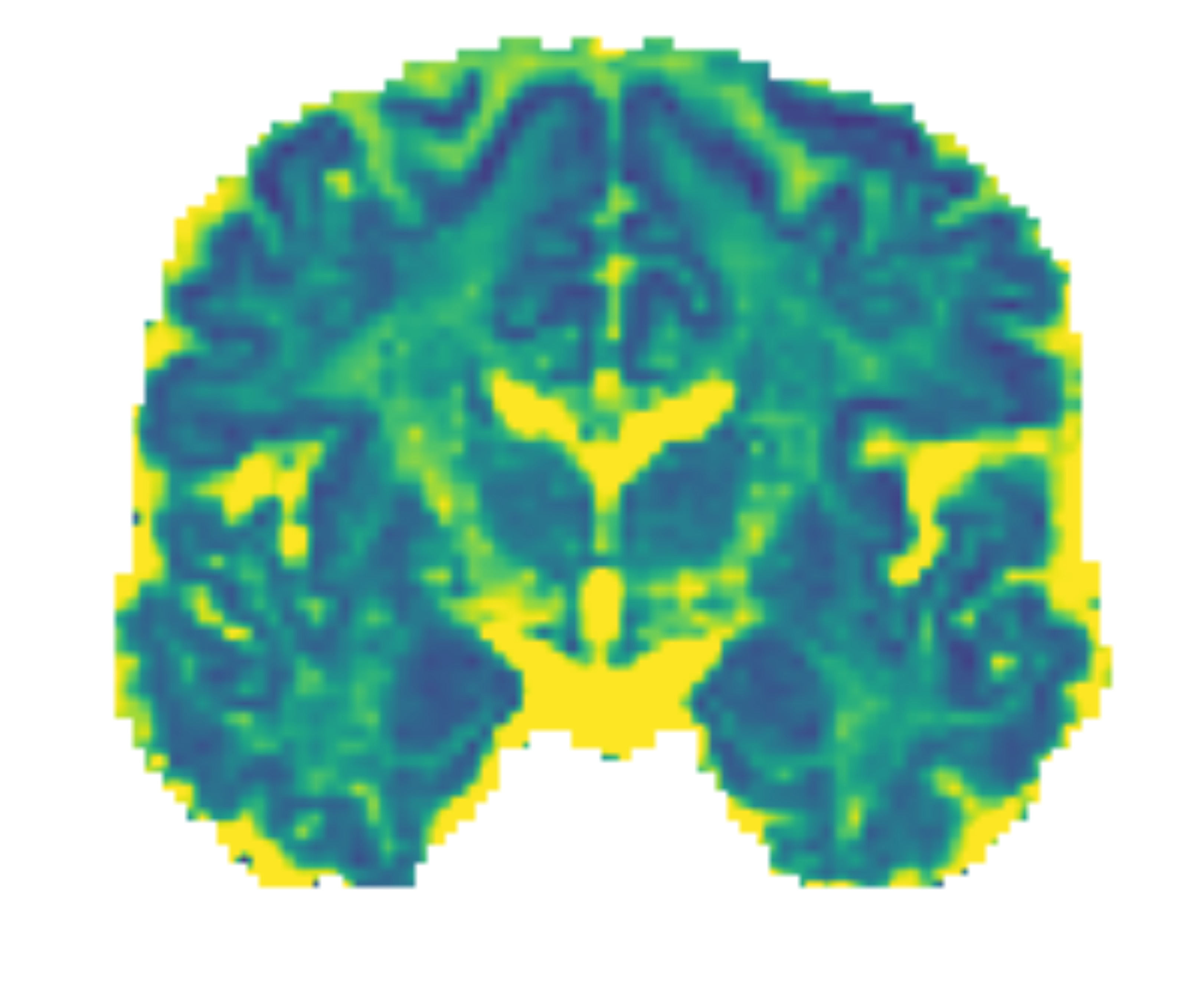
Sources:
Maëliss Jallais, Pedro Luiz Coelho Rodrigues, Alexandre Gramfort, Demian Wassermann. Inverting brain grey matter models with likelihood-free inference: a tool for trustable cytoarchitecture measurements, Journal of Machine Learning for Biomedical Imaging, pp.1-27, 2022
Powell, E; Battocchio, M; Parker, CS; Slator, PJ. Generalised Hierarchical Bayesian Microstructure Modelling for Diffusion MRI. CDMRI 2021 Proceedings. (pp. 36-47), 2021
Application: Uncovering the finer relationship between brain regions
As described in the parcellation application, functional Magnetic Resonance Imaging (fMRI) uncovers the function of different brain regions. However, current analysis often limits itself to correlation, that indicates the co-activation of brain regions but not their finer relationship. If two regions co-activate, is this because one activates the other, or the reverse or a third region activates both?
In this project, I apply PAVI to fit such dynamic models to fMRI signals and uncover the coupling between brain regions. The main hurdle is that fMRI measures the oxygenation of the blood in a given part of the brain, which is only an indirect proxy of the activity in that region. This so-called hemodynamic response function (HRF) is unknown and can vary across different brain regions. How can we consider this uncertainty when determining the coupling between different regions? Below is an example of the uncovered dynamical systems in a rat brain between the Motor cortex, the Insula and the Thalamus (figure from Ryiali et al. (2016)):
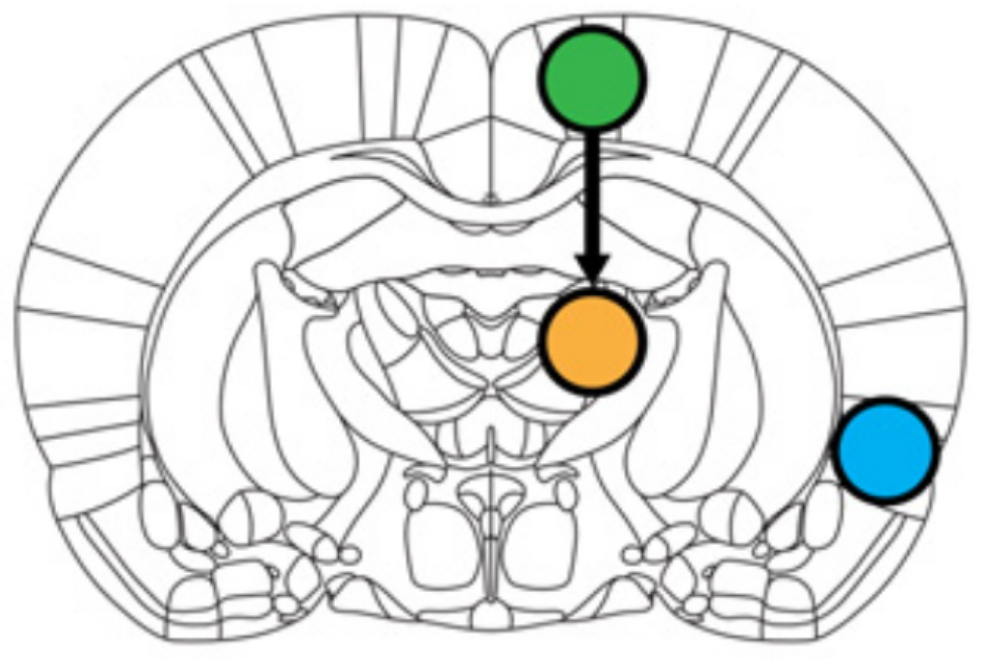
Sources:
Srikanth Ryali, Kaustubh Supekar, Tianwen Chen, Vinod Menon, Multivariate dynamical systems models for estimating causal interactions in fMRI, Neuroimage 54(2), 807-23, 2011
Srikanth Ryali ,Yen-Yu Ian Shih, Tianwen Chena, John Kochalka, Daniel Albaugh, Zhongnan Fang, Kaustubh Supekar, Jin Hyung Lee, Vinod Menon, Combining optogenetic stimulation and fMRI to validate a multivariate dynamical systems model for estimating causal brain interactions, NeuroImage 132, 398–405, 2016
